Playlist
Show Playlist
Hide Playlist
Ionization – Amino Acids
-
02 Basic AminoAcids.pdf
-
Reference List Biochemistry.pdf
-
Download Lecture Overview
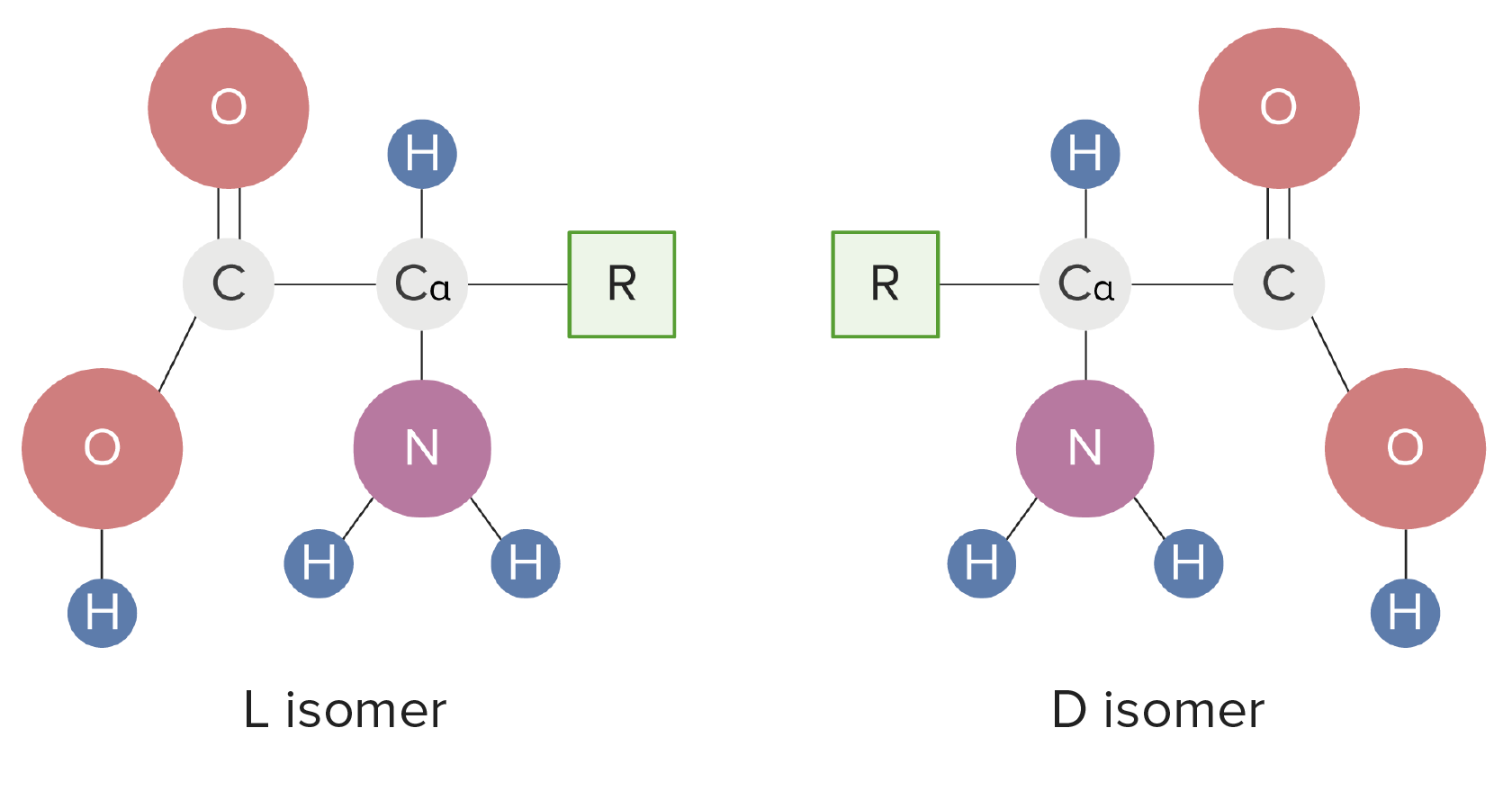
00:01 Now understanding amino acids and how they ionize is important and it's also a little complicated. So what I've done here as I've drawn on the screen, aspartic acid and the four different forms that it can exist as with ionization. On the left you see aspartic acid containing all of the protons that it can possibly hold, and on the right you see aspartic acid having lost all of the protons that it can lose. In between we see intermediate states. Now the ionization of the three groups within aspartic acid are all governed by pKa values. The lowest pKa value for an ionizable group in aspartic acid is that of the alpha carboxyl. The alpha carboxyl is about 2, the pKa value for the R group carboxyl is about four and that's labelled as pK2 on the diagram and the pKa for the alpha amine is about 9,5 and its labelled as pK3. 01:02 Let's step through the ionization scheme now for aspartic acid. What I've shown on the graph here is a plot of the pH of the solution compared to the number of equivalents of sodium hydroxide added. We can see if we start the graph that at the very lowest point we have a pH of zero. If we add a half equivalent of sodium hydroxide, what we do is we remove half of the protons of the first group with the lowest pKa. That means therefore, that if we analyze this that we have half of the molecules in the solution that will have the structure shown on the left with all the protons on, and half of the molecules in the solution that will have the structure on the right with the proton missing from the alpha carboxyl, and the protons on the other two groups unaffected. If we add an additional equivalent of sodium hydroxide to make a total of 1, we completely remove the first proton which was the one that was on the alpha carboxyl. But now we take an additional half of the protons off of the second group that ionizes, that of the R group carboxyl as shown here. In this case, half of the molecules have a structure shown on the left meaning that half of them have the proton off the alpha carboxyl, have the proton on the R group carboxyl and have the proton on the R group amine. The other half of the molecules have the structure shown on the right with a proton is missing from the alpha carboxyl, the R group carboxyl, but is still retained on the alpha amine. If we add one more equivalent of sodium hydroxide, we reach the pKa value shown here of about 9,5. Half of the aspartic acid molecules will have the structure that's shown on the left, that is, protons missing from the alpha carboxyl and the R group carboxyl but retained on the alpha amine. The other half of the molecules will have protons missing from all of the groups. The overall charge of the molecule on the right is -2. 03:08 Now if we're interested in understanding what the charges on molecules are, we can't really compare them at half equivalents because we have mixtures of the two, but if we used full equivalents for removing of protons than we can do that. 03:25 Let's analyze the charge then of aspartic acid as a function of pH moving through the solution. At the very lowest pH that we started out was a pH of 0. A pH of 0 is 2,2 pH units below the first pKa. When we assign charges to amino acids, we can make reasonable assumptions that if we're one or more pH units below the pKa value for an ionizable group, that that ionizable group will contain all of its protons. In this case the ionizable group that would lose its protons first is the alpha carboxyl, but because we're more than one pH unit below its pKa value, it retains its proton. Zero is of course far below the pKa values of the other two ionizable groups and so they also retain their protons. Adding up the charges here, the molecule has an overall charge of +1. If we add one equivalent of sodium hydroxide, we completely remove the first ionizable proton, that corresponding to the one on the alpha carboxyl group. This gives us the structure that we see on the screen, the structure that we see on the screen lacks a proton, the alpha carboxyl, has protons on the alpha amine and the R group amine. The overall charge of this molecule is 0. Now amino acids that have a charge of 0 are called zwitterions. 04:43 You'll also notice on the graph that this pH is marked as pI and the pI has a special name because it's a special pH. The pI is the pH at which a molecule has a charge of exactly 0. A zwitterion will always exist at the pI value for an amino acid. 05:05 If we add another equivalent of sodium hydroxide, we remove the proton completely from the R group carboxyl and we're left with the structure that's seen on the screen. Protons missing from the R group and the alpha carboxyl but retained by the alpha amine. If we add one more equivalent of sodium hydroxide, we've removed the last of the protons and create a molecule now that has a charge of -2. Protons missing from all the groups 0 charge on the alpha amine and minus charges on the two carboxyls. 05:37 This table has a lot of information on it and I don't show it to you to overwhelm you with information, but rather to show you some of the things that we know about amino acids. 05:46 The first column has the name of the amino acid and the second column contains information about the three letter abbreviation we commonly use to designate amino acids. One letter codes are used for even more compacting of the sequence information contained in the third column. 06:02 The fourth column describes the polarity, and the fifth column describes the charges that can exist at physiological pH as I have described in the lecture. The next column is interesting in that it shows the hydropathy index. The hydropathy index is a measure of the tendency of the R group of the amino acid to associate with water. More positive values mean a lower tendency to interact with water, whereas more negative values indicate a greater tendency to associate with water. Some R groups of amino acids absorb ultraviolet light, this is shown in the seventh column under absorbance. This indicates the wavelength of the ultraviolet light that's absorbed by individual R groups of some of the amino acids. Epsilon is plotted in column number eight and epsilon is the absorptivity coefficient that corresponds to the absorbance of light. In the last column we have the plot of the molecular weight for each amino acid and you can see that each amino acid has an average molecular weight overall of about 110 to 120. This concludes the description of the basic structures, features and properties of the 20 amino acids found in proteins.
About the Lecture
The lecture Ionization – Amino Acids by Kevin Ahern, PhD is from the course Biochemistry: Basics.
Included Quiz Questions
Which of the following amino acid is positively charged at physiological pH?
- Arginine
- Isoleucine
- Valine
- Asparagine
- Glycine
The hydropathy index is a measure of which of the following?
- The tendency of the R group of an amino acid to associate with water
- The tendency of the -NH2 group of an amino acid to associate with water
- The tendency of the R group of an amino acid to associate with other amino acids
- The tendency of the -COOH group of an amino acid to associate with water
- The tendency of the -NH2 and -COOH groups of an amino acid to associate with other macromolecules
The isoelectric point is located at what pH?
- The pH at which an amino acid carries no net electrical charge on it
- The pH at which an amino acid carries net positive charge on it
- The pH at which an amino acid carries net negative charge on it
- The pH at which an amino acid carries extra protons on it
- The pH at which an amino acid denotes all the protons present in it
Customer reviews
5,0 of 5 stars
| 5 Stars |
|
1 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |
I loved this lecture ´cause dr ahern was so clear and it really helped me understand this topic.